Introduction to PhasorPy#
An introduction to using the PhasorPy library.
PhasorPy is an open-source Python library for the analysis of fluorescence lifetime and hyperspectral images using the Phasor approach.
Using the PhasorPy library requires familiarity with the phasor approach, image processing, array programming, and Python.
Install Python#
An installation of Python version 3.11 or higher is required to use the PhasorPy library. Python is an easy to learn, powerful programming language. Python installers can be obtained from, for example, Python.org or Anaconda.com. Alternatively, Python can be used via Google Colab, a free, cloud-based service. Refer to the Python Tutorial for an introduction to Python.
Install PhasorPy#
To download and install the PhasorPy library and all its dependencies from the Python Package Index (PyPI), run the following command on a command prompt, shell, or terminal:
python -m pip install -U "phasorpy[all]"
Alternatively, PhasorPy can be installed via conda-forge in an Anaconda environment:
conda install conda-forge::phasorpy
The development version of PhasorPy can be installed instead from the latest source code on GitHub. This requires a C compiler, such as XCode, Visual Studio, or gcc, to be installed:
python -m pip install -U git+https://github.com/phasorpy/phasorpy.git
Update optional dependencies as needed:
python -m pip install -U lfdfiles sdtfile ptufile
Import phasorpy#
Start the Python interpreter, import the phasorpy package,
and print its version:
import phasorpy
print(phasorpy.__version__)
0.6
Besides the PhasorPy library, the numpy library is used for array computing throughout this tutorial:
import numpy
Read signal from file#
The phasorpy.io module provides functions to read time-resolved
and hyperspectral image stacks and metadata from many file formats used
in microscopy, for example PicoQuant PTU, OME-TIFF, Zeiss LSM, and files
written by SimFCS software.
However, any other means that yields image stacks in numpy-array compatible
form can be used instead.
Image stacks, which may have any number of dimensions, are referred to as
signal in the PhasorPy library.
The phasorpy.datasets module provides access to various sample
files. For example, an ImSpector TIFF file from the
FLUTE project containing a
time-correlated single photon counting (TCSPC) histogram
of a zebrafish embryo at day 3, acquired at 80.11 MHz:
from phasorpy.datasets import fetch
from phasorpy.io import signal_from_imspector_tiff
signal = signal_from_imspector_tiff(fetch('Embryo.tif'))
frequency = signal.attrs['frequency']
print(signal.shape, signal.dtype)
(56, 512, 512) uint16
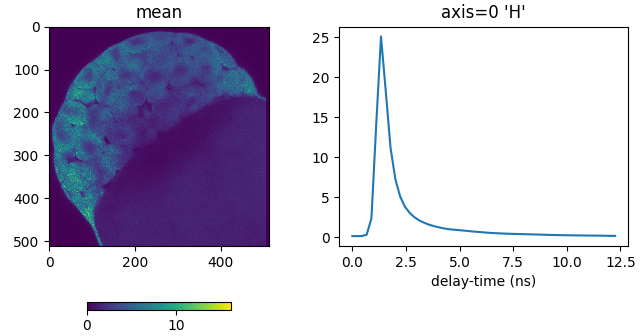
Plot the spatial and histogram averages. The histogram bins are in the first dimension of the signal array (axis=’H’ or axis=0):
from phasorpy.plot import plot_signal_image
plot_signal_image(signal, axis='H', xlabel='delay-time (ns)')
Calculate phasor coordinates#
The phasorpy.phasor module provides functions to calculate,
calibrate, filter, and convert phasor coordinates.
Phasor coordinates are the real and imaginary components of the complex
numbers returned by a real forward Digital Fourier Transform (DFT)
of a signal at certain harmonics (multiples of the fundamental frequency),
normalized by the mean intensity (the zeroth harmonic).
Phasor coordinates are named real and imag in the PhasorPy library.
In literature and other software, they are also known as
\(G\) and \(S\) or \(a\) and \(b\) (as in \(a + bi\)).
Phasor coordinates of the first harmonic are calculated from the signal over the axis containing the TCSPC histogram bins (axis=’H’ or axis=0):
from phasorpy.phasor import phasor_from_signal
mean, real, imag = phasor_from_signal(signal, axis='H')
The phasor coordinates are undefined if the mean intensity is zero.
In that case, the arrays contain special NaN (Not a Number) values,
which are ignored in further analysis:
print(real[:4, :4])
[[ nan nan -0.27292411 nan]
[ nan nan 0.16095963 nan]
[ nan nan 0.90096887 nan]
[ nan 0.6234898 nan nan]]
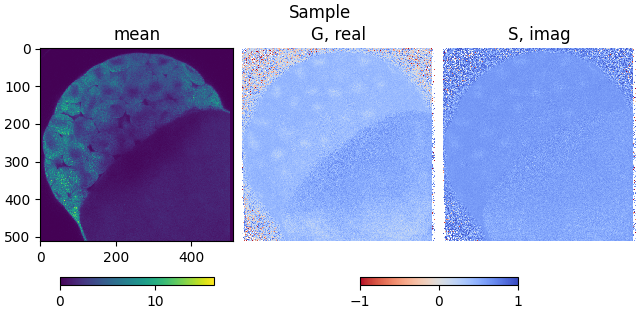
Plot the calculated phasor coordinates:
from phasorpy.plot import plot_phasor_image
plot_phasor_image(mean, real, imag, title='Sample')
By default, only the phasor coordinates at the first harmonic are calculated. However, only when the phasor coordinates at all harmonics are considered (including the mean intensity) is the signal completely described:
from phasorpy.phasor import phasor_to_signal
phasor_all_harmonics = phasor_from_signal(signal, axis=0, harmonic='all')
reconstructed_signal = phasor_to_signal(
*phasor_all_harmonics, axis=0, samples=signal.shape[0]
)
numpy.testing.assert_allclose(
numpy.nan_to_num(reconstructed_signal), signal, atol=1e-3
)
Calibrate phasor coordinates#
The signals from time-resolved measurements are convoluted with an instrument response function, causing the phasor-coordinates to be phase-shifted and modulated (scaled) by unknown amounts. The phasor coordinates must therefore be calibrated with coordinates obtained from a reference standard of known lifetime, acquired with the same instrument settings.
In this case, a homogeneous solution of Fluorescein with a lifetime of 4.2 ns was imaged.
Read the signal of the reference measurement from a file:
reference_signal = signal_from_imspector_tiff(fetch('Fluorescein_Embryo.tif'))
assert reference_signal.attrs['frequency'] == frequency
Calculate phasor coordinates from the measured reference signal:
reference_mean, reference_real, reference_imag = phasor_from_signal(
reference_signal, axis=0
)
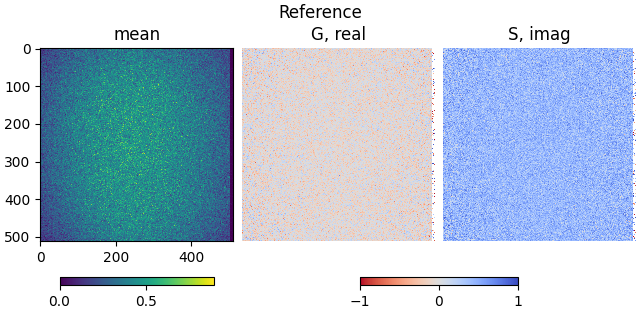
Show the calculated reference phasor coordinates:
plot_phasor_image(
reference_mean, reference_real, reference_imag, title='Reference'
)
Calibrate the raw phasor coordinates with the reference coordinates of known lifetime (Fluorescein, 4.2 ns):
from phasorpy.phasor import phasor_calibrate
real, imag = phasor_calibrate(
real,
imag,
reference_mean,
reference_real,
reference_imag,
frequency=frequency,
lifetime=4.2,
)
Show the calibrated phasor coordinates:
plot_phasor_image(mean, real, imag, title='Calibrated')
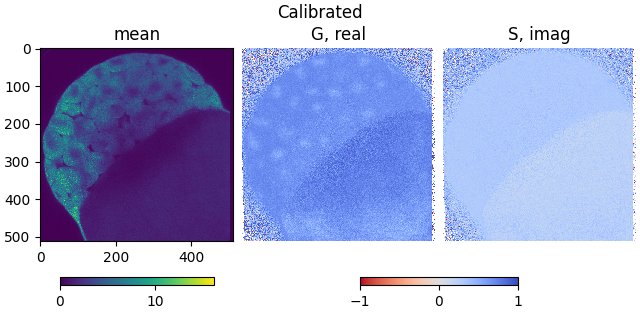
The phasor coordinates are now located in the first quadrant, except for some with low signal to noise level.
If necessary, the calibration can be undone/reversed using the same reference:
uncalibrated_real, uncalibrated_imag = phasor_calibrate(
real,
imag,
reference_mean,
reference_real,
reference_imag,
frequency=frequency,
lifetime=4.2,
reverse=True,
)
numpy.testing.assert_allclose(
(mean, uncalibrated_real, uncalibrated_imag),
phasor_from_signal(signal, axis=0),
atol=1e-3,
)
Filter phasor coordinates#
Applying median filter to the calibrated phasor coordinates, often multiple times, improves contrast and reduces noise. The filter is applied independently to the real and imaginary components, but not to the signal average:
from phasorpy.phasor import phasor_filter_median
mean, real, imag = phasor_filter_median(mean, real, imag, size=3, repeat=2)
Pixels with low intensities are commonly excluded from analysis and visualization of phasor coordinates:
from phasorpy.phasor import phasor_threshold
mean, real, imag = phasor_threshold(mean, real, imag, mean_min=1)
Show the calibrated, filtered phasor coordinates:
plot_phasor_image(
mean, real, imag, title='Calibrated, filtered phasor coordinates'
)
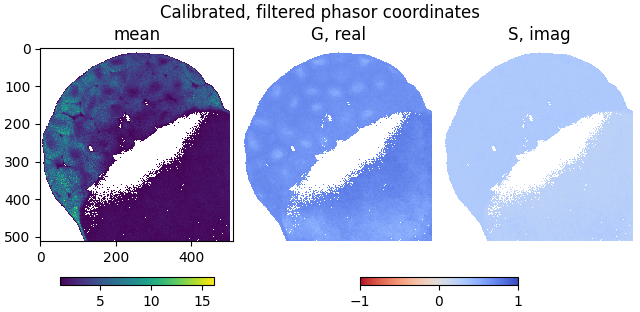
Store phasor coordinates#
Phasor coordinates and select metadata can be exported to OME-TIFF formatted files, which are compatible with Bio-Formats and Fiji.
Write the calibrated and filtered phasor coordinates, and frequency to an OME-TIFF file:
from phasorpy.io import phasor_from_ometiff, phasor_to_ometiff
phasor_to_ometiff(
'phasors.ome.tif',
mean,
real,
imag,
frequency=frequency,
harmonic=1,
description=(
'Phasor coordinates of a zebrafish embryo at day 3, '
'calibrated, median-filtered, and thresholded.'
),
)
Read the phasor coordinates and metadata back from the OME-TIFF file:
mean_, real_, imag_, attrs = phasor_from_ometiff('phasors.ome.tif')
numpy.allclose(real_, real)
assert real_.dtype == numpy.float32
assert attrs['frequency'] == frequency
assert attrs['harmonic'] == 1
assert attrs['description'].startswith('Phasor coordinates of')
These functions also work transparently with multi-harmonic phasor coordinates.
Plot phasor coordinates#
The phasorpy.plot module provides functions and classes for
plotting phasor and polar coordinates.
Large number of phasor coordinates, such as obtained from imaging, are commonly visualized as two-dimensional histograms:
from phasorpy.plot import plot_phasor
plot_phasor(
real,
imag,
frequency=frequency,
title='Calibrated, filtered phasor coordinates',
)
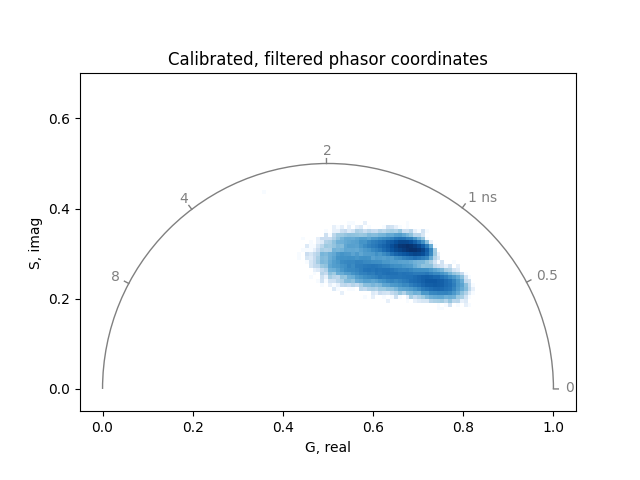
The calibrated phasor coordinates of all pixels lie inside the universal semicircle (on which theoretically the phasor coordinates of all single exponential lifetimes are located). That means, all pixels contain mixtures of signals from multiple lifetime components.
For comparison, the uncalibrated, unfiltered phasor coordinates:
plot_phasor(
uncalibrated_real,
uncalibrated_imag,
allquadrants=True,
title='Raw phasor coordinates',
)
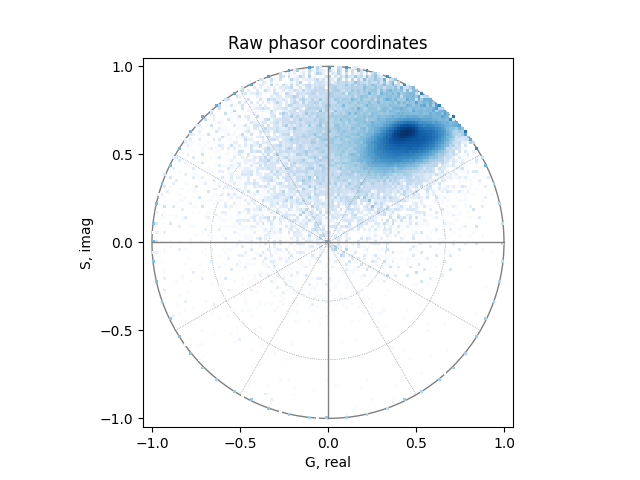
Select phasor coordinates#
The phasorpy.cursors module provides functions for selecting phasor
coordinates to define and mask regions of interest within the phasor space.
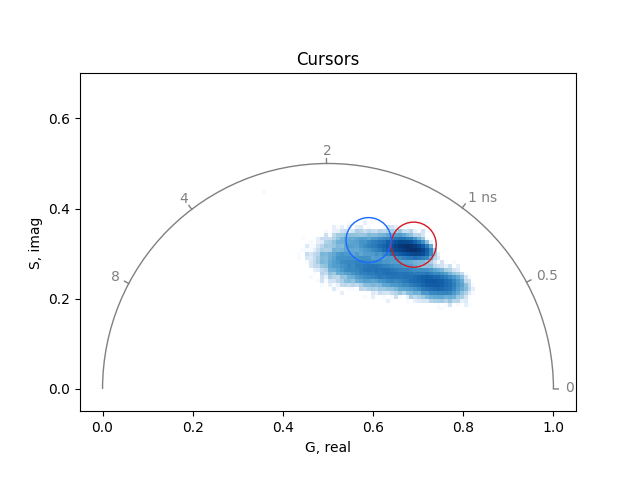
Mask regions of interest in the phasor space using circular cursors:
from phasorpy.color import CATEGORICAL
from phasorpy.cursors import mask_from_circular_cursor
cursor_real = 0.69, 0.59
cursor_imag = 0.32, 0.33
radius = 0.05, 0.05
cursors_masks = mask_from_circular_cursor(
real, imag, cursor_real, cursor_imag, radius=radius
)
Plot the cursors in distinct colors using the higher-level
PhasorPlot interface:
from phasorpy.plot import PhasorPlot
phasorplot = PhasorPlot(frequency=frequency, title='Cursors')
phasorplot.hist2d(real, imag)
for i in range(len(cursor_real)):
phasorplot.circle(
cursor_real[i],
cursor_imag[i],
radius=radius[i],
color=CATEGORICAL[i],
linestyle='-',
)
phasorplot.show()
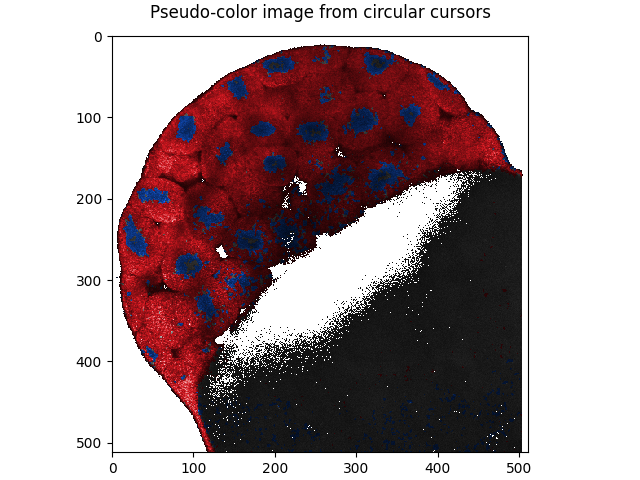
Blend the cursor masks with the mean intensity image to produce a pseudo-colored image:
from phasorpy.cursors import pseudo_color
from phasorpy.plot import plot_image
pseudo_color_image = pseudo_color(*cursors_masks, intensity=mean)
plot_image(
pseudo_color_image, title='Pseudo-color image from circular cursors'
)
Component analysis#
# TODO
Spectral phasors#
Phasor coordinates can be calculated from hyperspectral images (acquired at many equidistant emission wavelengths) and processed in much the same way as time-resolved signals. Calibration is not necessary.
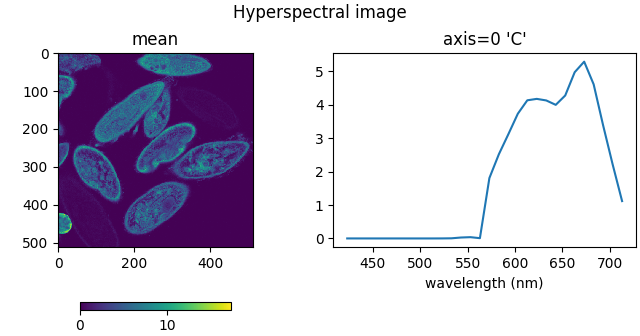
Open a hyperspectral dataset acquired with a laser scanning microscope at 30 emission wavelengths:
from phasorpy.io import signal_from_lsm
hyperspectral_signal = signal_from_lsm(fetch('paramecium.lsm'))
plot_signal_image(
hyperspectral_signal,
axis=0,
title='Hyperspectral image',
xlabel='wavelength (nm)',
)
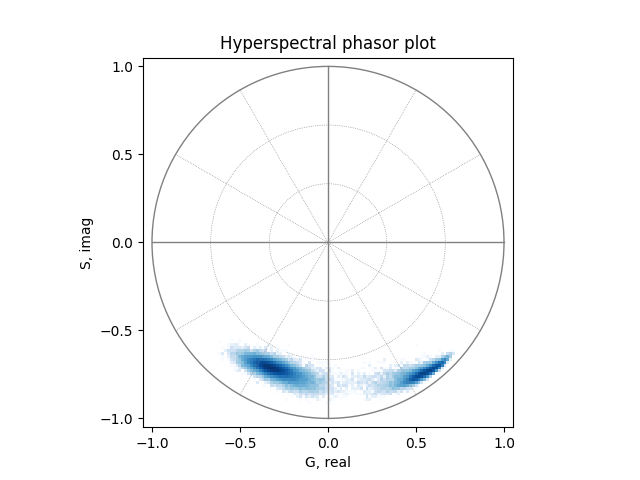
Calculate phasor coordinates at the first harmonic and filter out pixels with low intensities:
mean, real, imag = phasor_from_signal(hyperspectral_signal, axis=0)
_, real, imag = phasor_threshold(mean, real, imag, mean_min=1)
Plot the phasor coordinates as a two-dimensional histogram:
plot_phasor(real, imag, allquadrants=True, title='Hyperspectral phasor plot')
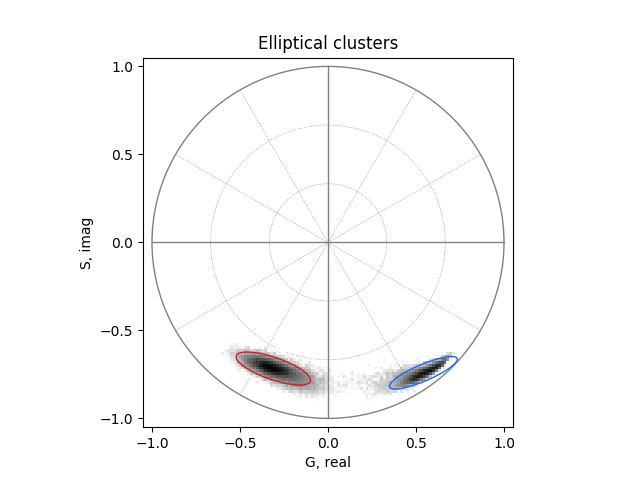
Find clusters#
Automatically find the two elliptical clusters in the phasor space using a Gaussian mixture model and plot them in distinct colors:
from phasorpy.cluster import phasor_cluster_gmm
center_real, center_imag, radius, radius_minor, angle = phasor_cluster_gmm(
real, imag, clusters=2
)
phasorplot = PhasorPlot(allquadrants=True, title='Elliptical clusters')
phasorplot.hist2d(real, imag, cmap='Greys')
for i in range(len(center_real)):
phasorplot.cursor(
center_real[i],
center_imag[i],
radius=radius[i],
radius_minor=radius_minor[i],
angle=angle[i],
color=CATEGORICAL[i],
linestyle='-',
)
phasorplot.show()
Use the elliptic clusters to mask regions of interest in the phasor space:
from phasorpy.cursors import mask_from_elliptic_cursor
elliptic_masks = mask_from_elliptic_cursor(
real,
imag,
center_real,
center_imag,
radius=radius,
radius_minor=radius_minor,
angle=angle,
)
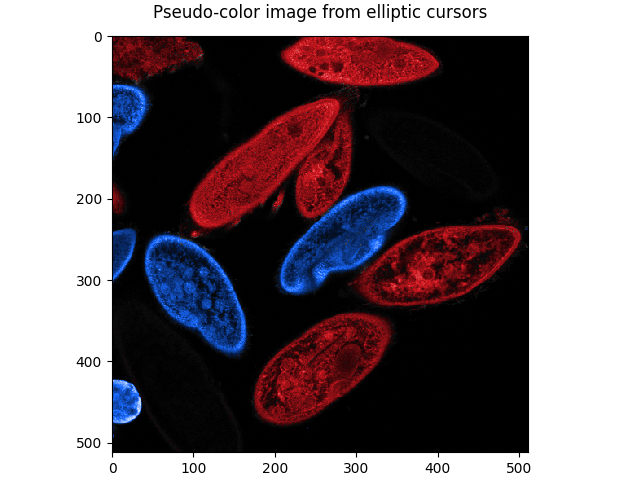
Plot a pseudo-color image, composited from the elliptic cursor masks and the mean intensity image:
pseudo_color_image = pseudo_color(*elliptic_masks, intensity=mean)
plot_image(
pseudo_color_image, title='Pseudo-color image from elliptic cursors'
)
Appendix#
Print information about Python interpreter and installed packages:
from phasorpy.utils import versions
print(versions())
Python-3.12.10
phasorpy-0.6
numpy-2.3.1
tifffile-2025.6.11
imagecodecs-n/a
lfdfiles-2025.5.10
sdtfile-2025.5.10
ptufile-2025.5.10
liffile-2025.5.10
matplotlib-3.10.3
scipy-1.16.0
skimage-n/a
sklearn-1.7.0
pandas-2.3.0
xarray-2025.6.1
click-8.2.1
pooch-1.8.2
sphinx_gallery_thumbnail_number = -8 mypy: allow-untyped-defs, allow-untyped-calls mypy: disable-error-code=”arg-type, assignment” isort: skip_file
Total running time of the script: (0 minutes 10.848 seconds)